Table of Contents
Introduction:
- Sanger sequencing, also known as dideoxy sequencing, is a method of DNA sequencing that was developed by Frederick Sanger in 1977.
- It involves adding a short strand of DNA that is complementary to the target DNA molecule, along with four different dideoxynucleotides (ddNTPs), which are building blocks that stop DNA synthesis when incorporated into the growing DNA strand.
- By repeating the process several times and using different ddNTPs each time, a series of fragments of varying lengths are produced. These fragments are then separated by size using gel electrophoresis and read to determine the order of the nucleotides in the target DNA molecule.
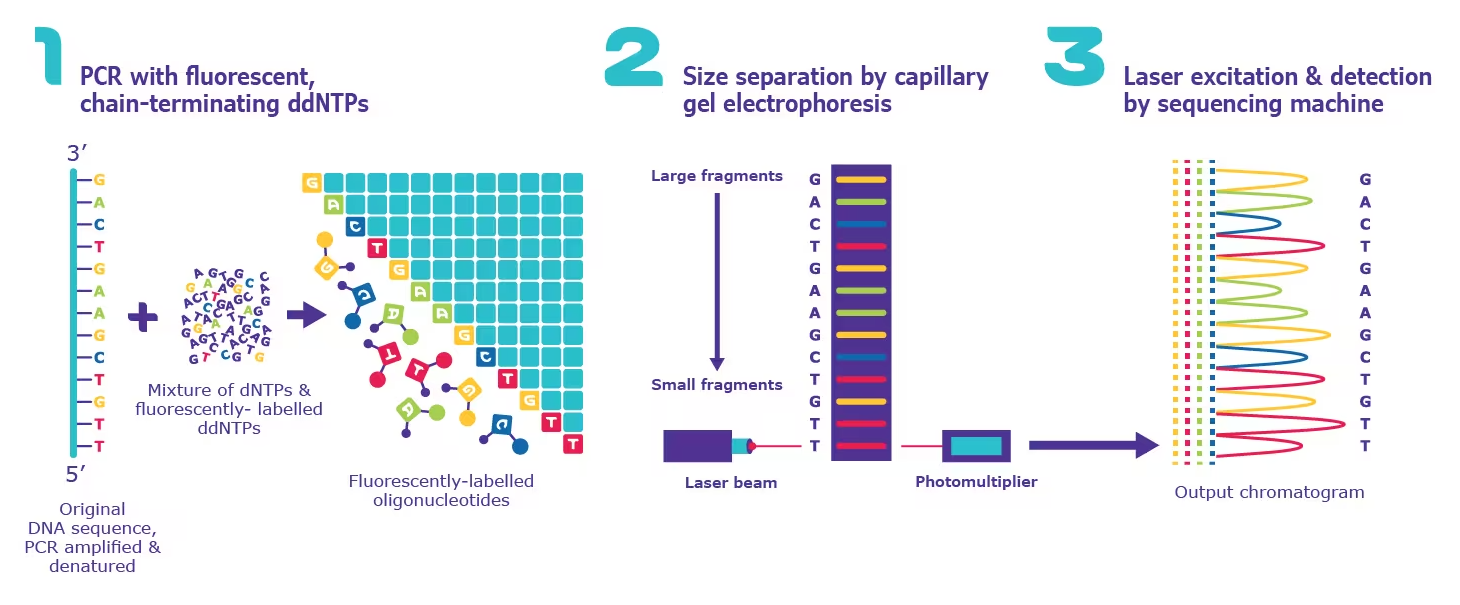
Principle of Sanger Sequencing:
- Sanger sequencing works by using a DNA polymerase, such as Taq polymerase, to synthesize a new DNA strand complementary to the target DNA molecule.
- During the synthesis process, a small amount of each of the four different ddNTPs is added to the reaction. These ddNTPs are similar to the normal nucleotides used for DNA synthesis (dNTPs), but they contain a different chemical structure that stops DNA synthesis when incorporated into the growing DNA strand.
- The result is a series of DNA fragments of varying lengths, each with a different ddNTP incorporated into its end.
Steps of Sanger Sequencing:
- Preparation of Template DNA:
- The target DNA molecule is prepared for sequencing by extracting it from the source material and purifying it to remove any contaminants.
- The DNA can then be amplified, if necessary, using PCR (Polymerase Chain Reaction) to generate enough material for sequencing.
- Primer Annealing:
- A short, single-stranded DNA primer is annealed to the template DNA, marking the starting point for DNA synthesis.
- Synthesis of Complementary Strand:
- DNA synthesis is initiated using a DNA polymerase, such as Taq polymerase, and a mixture of the four different ddNTPs and normal dNTPs.
- As the DNA polymerase extends the primer, it incorporates the ddNTPs into the growing DNA strand.
- When a ddNTP is incorporated, DNA synthesis stops, resulting in a series of DNA fragments of varying lengths.
- Gel Electrophoresis:
- The DNA fragments are separated by size using gel electrophoresis, a process that uses an electric field to separate DNA fragments based on their size and charge.
- Detection and Analysis:
- The separated DNA fragments are then transferred to a solid support, such as a nitrocellulose or nylon membrane, and exposed to a radioactive or fluorescent label.
- The label allows the fragments to be visualized and detected, and the sequence can be determined by reading the order of the incorporated ddNTPs.
Advantages of Sanger Sequencing:
- Sanger sequencing is a reliable and widely used method for small-scale DNA sequencing projects.
- It is capable of producing high-quality, high-resolution data, with a typical error rate of less than 1 in 10,000 nucleotides.
- Sanger sequencing is also widely used for sequencing individual genes and for validation of next-generation sequencing data.
Limitations of Sanger Sequencing:
- Sanger sequencing is a relatively slow and expensive method, and is not well-suited for large-scale sequencing projects.
- It requires a large amount of starting material, which can limit its usefulness for sequencing rare or degraded DNA.
- The maximum read length is limited to around 1000-2000 nucleotides, making it difficult to sequence large genomic regions or entire genomes.
Applications of Sanger Sequencing:
- Sanger sequencing has a wide range of applications, including:
- DNA sequencing for basic research and genome analysis, such as sequencing individual genes, genomic regions, and whole genomes.
- Validation of next-generation sequencing data, as Sanger sequencing can be used to confirm the accuracy of large-scale sequencing results.
- Medical testing, such as genetic testing for inherited diseases and cancer diagnosis.
- Identification of mutations and variations in DNA, such as single nucleotide polymorphisms (SNPs) and insertions/deletions (indels).
- Studying evolutionary relationships, as Sanger sequencing can be used to compare the DNA sequences of different species to determine their evolutionary relationships.
Conclusion:
- Sanger sequencing is a widely used and reliable method for DNA sequencing that has been essential for many important discoveries in the fields of genetics and genomics.
- Although it has some limitations, its high accuracy, long history of use, and widespread availability make it a valuable tool for a wide range of applications, from basic research to medical testing.